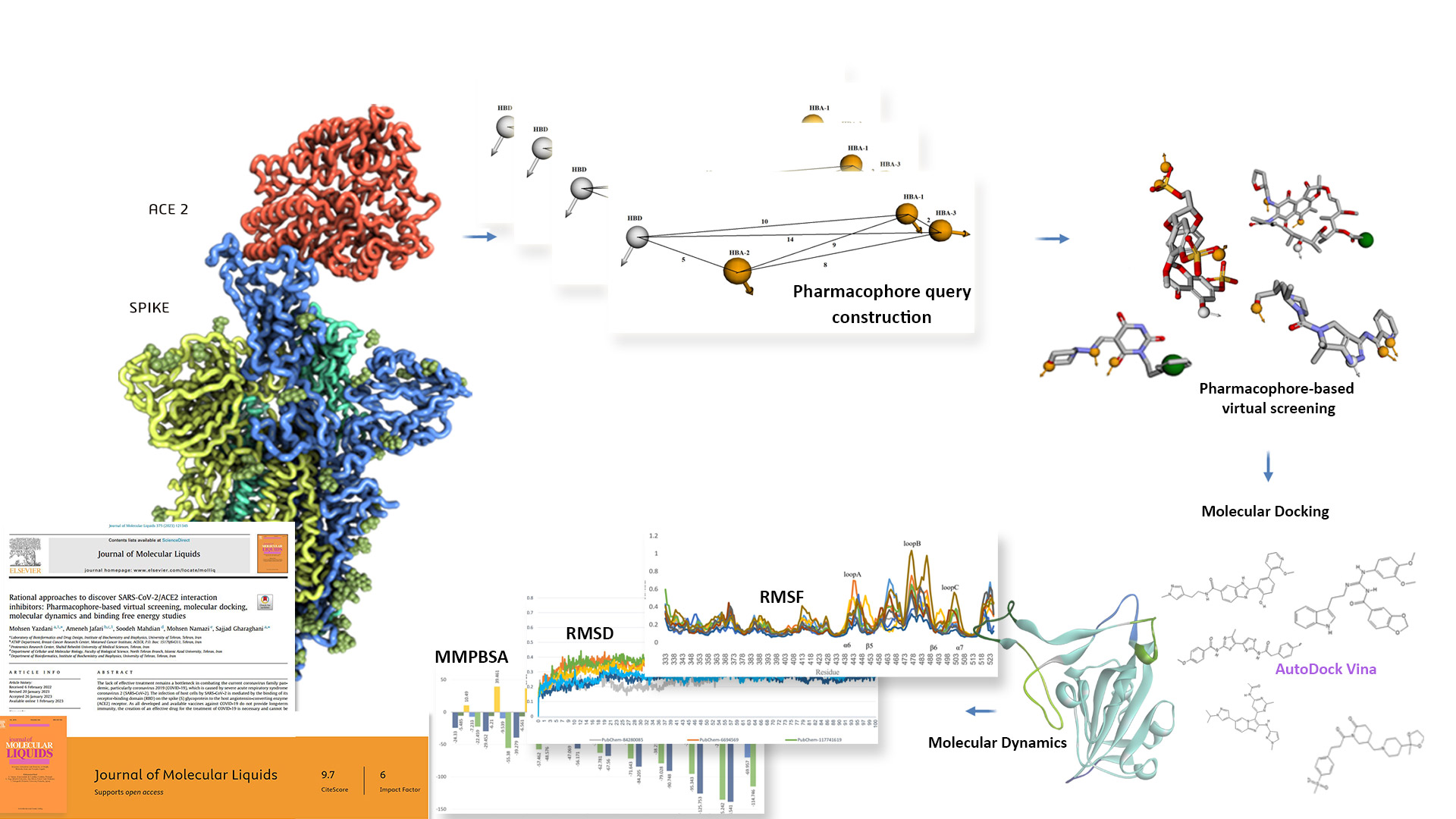
The project aims to discover potential inhibitors of the SARS-CoV-2 virus, which causes COVID-19, by using computational methods. The project uses the interaction between the virus’s spike protein and the human ACE2 receptor as a target for drug design. The project involves the following steps:
- Analyzing the structure and interface of the spike protein and the ACE2 receptor
- Constructing pharmacophore models based on the hot spot regions of the interaction
- Performing pharmacophore-based virtual screening on large compound databases
- Performing molecular docking, molecular dynamics, and binding free energy calculations on the selected compounds
- Identifying the most potent compounds that can block the interaction and disrupt the viral entry
The project is a novel and innovative approach to finding potential drugs for COVID-19 treatment. The project uses rational drug design techniques and advanced computational tools to screen millions of compounds and select the best candidates. The project also provides insights into the molecular mechanism of the interaction and the binding patterns of the inhibitors. The project can be applied to new variants of the virus and other viral targets. The project is a valuable contribution to the global effort to combat the COVID-19 pandemic.








