-
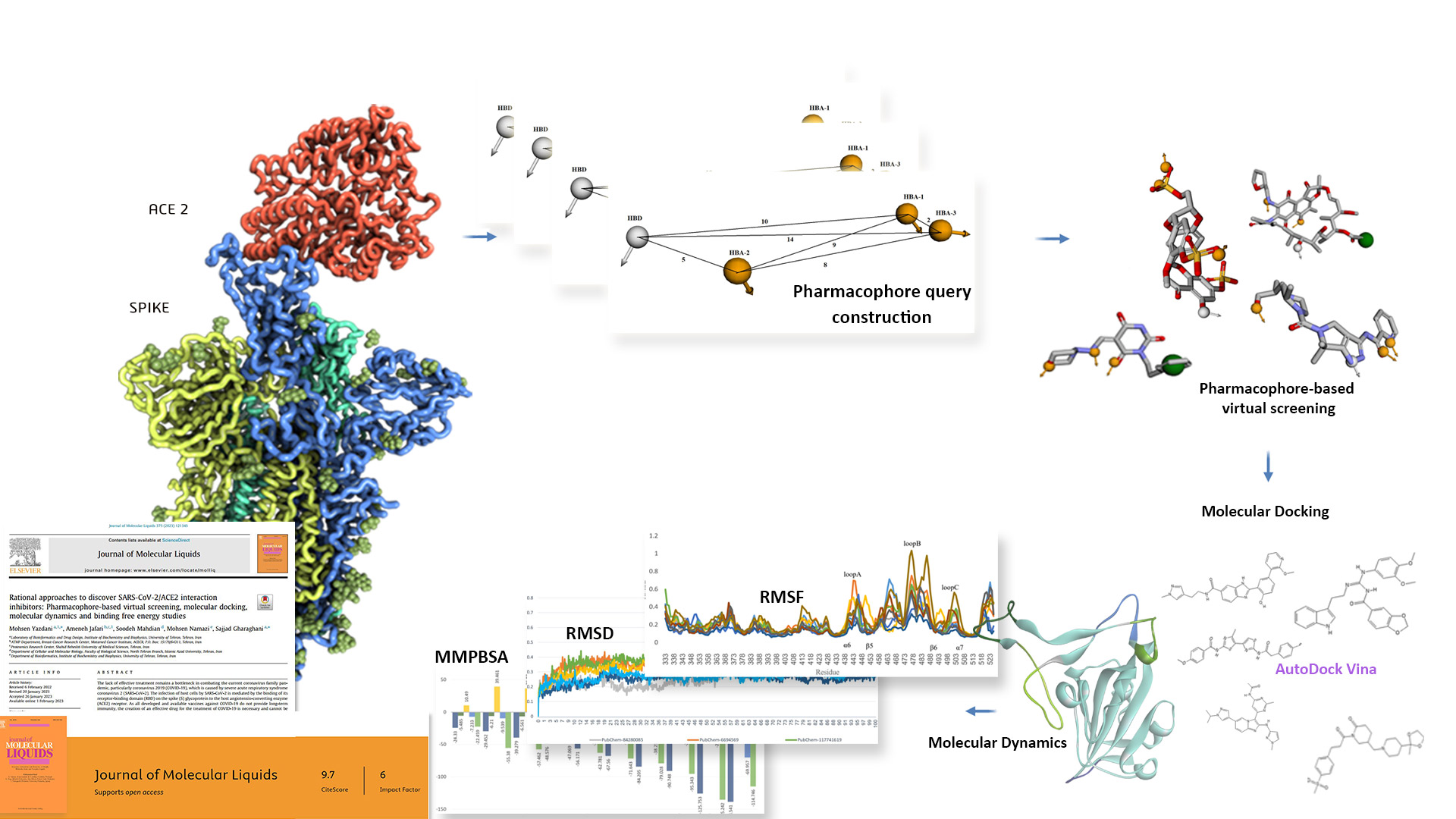
Rational approaches,
SARS-CoV-2/ACE2 PPI inhibitors -
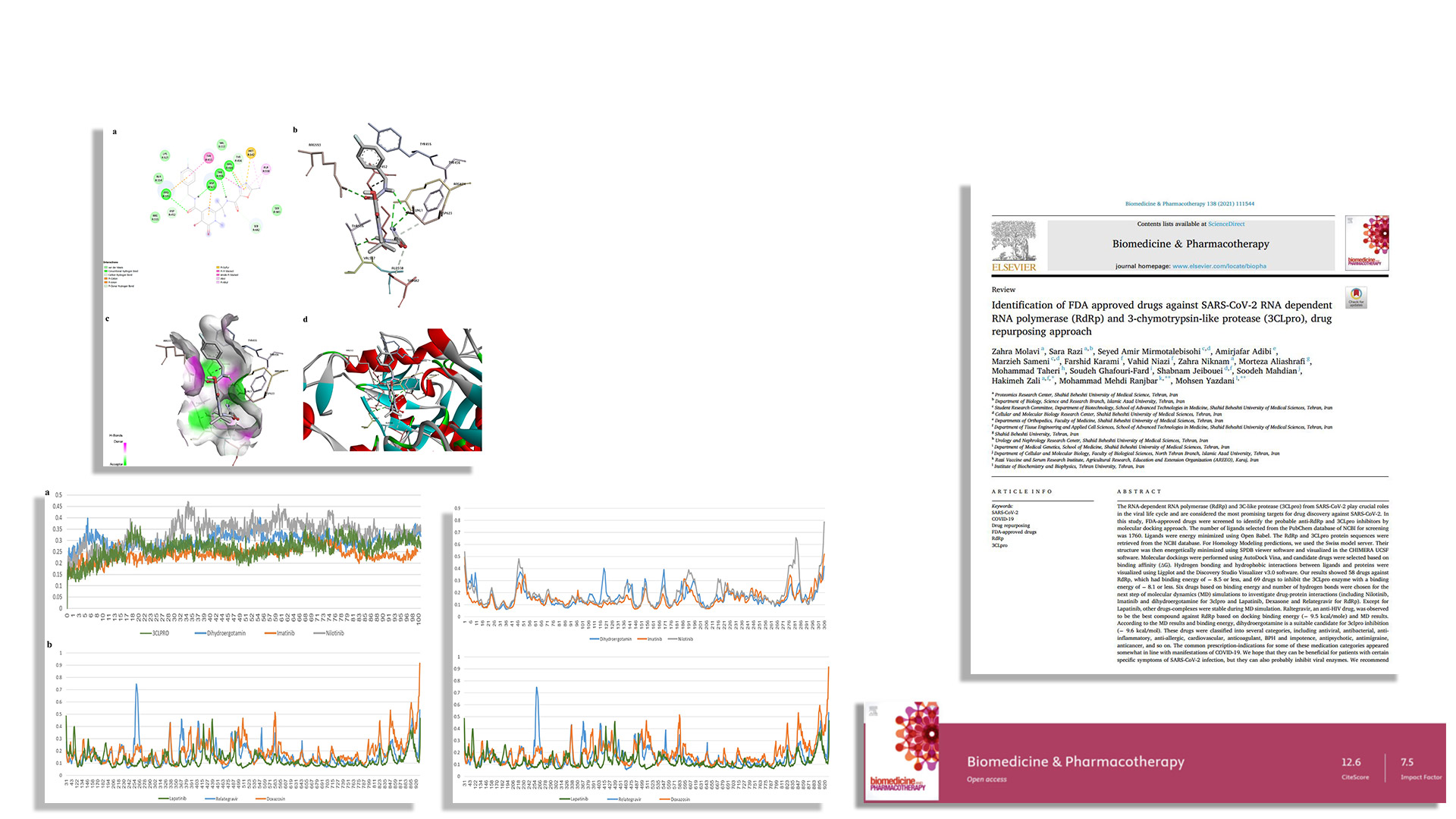
Drug repurposing approach,
FDA approved drugs against RdRp and 3CLpro. -
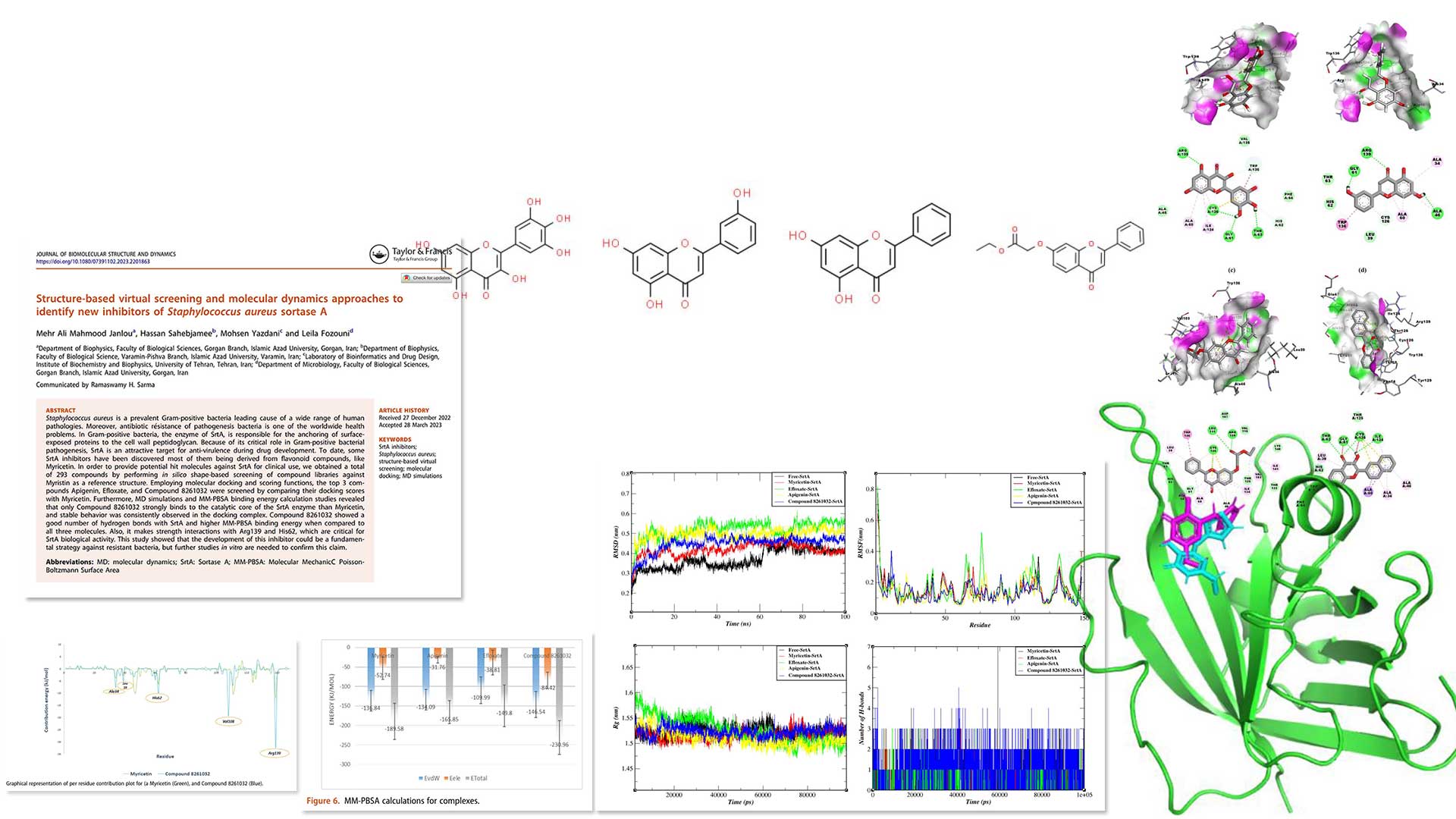
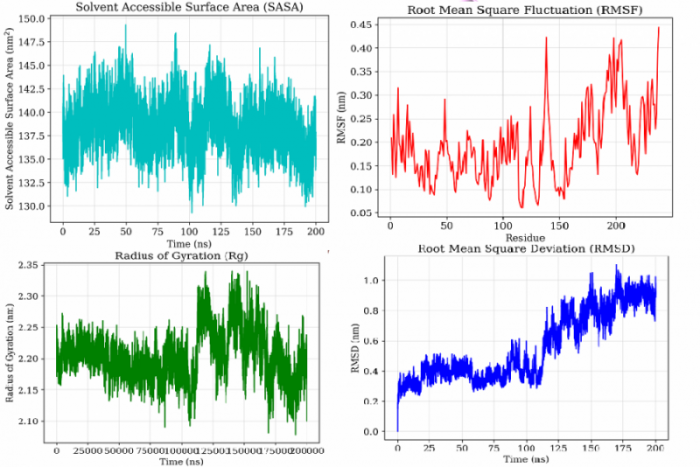
Structure-based virtual screening & MD,
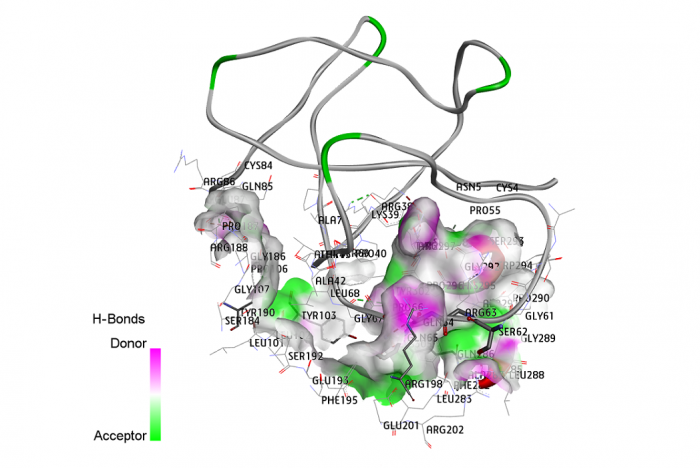
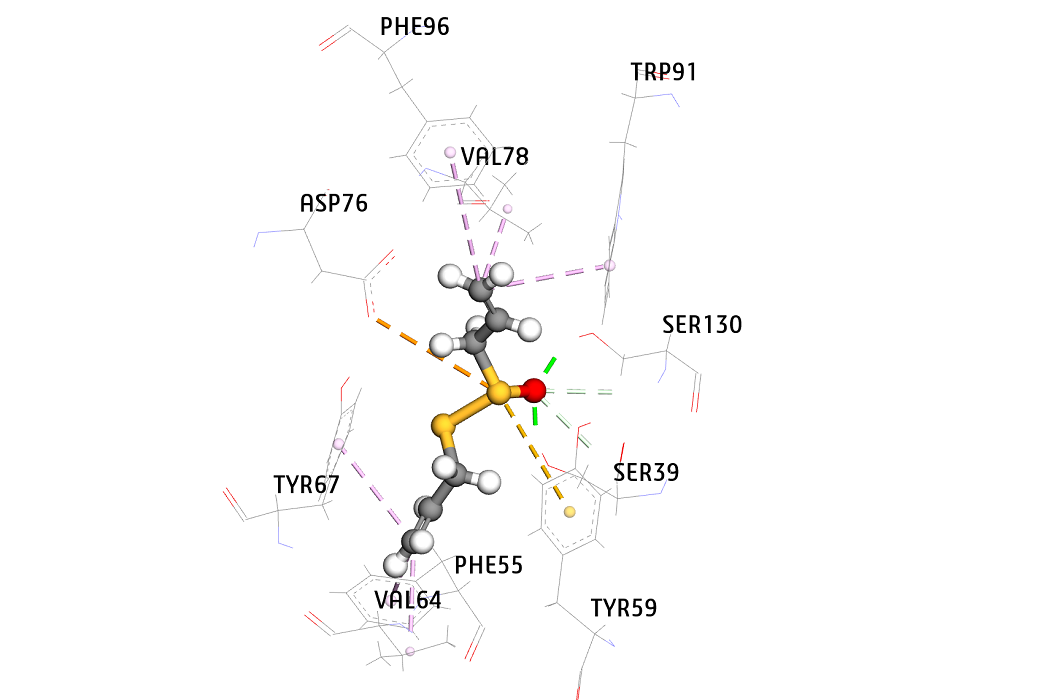
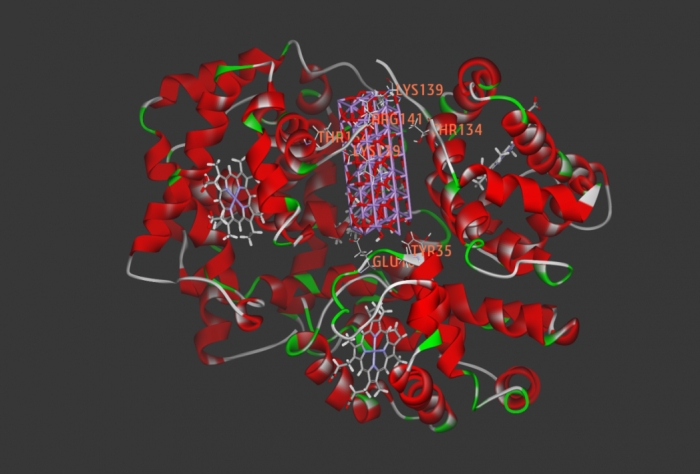
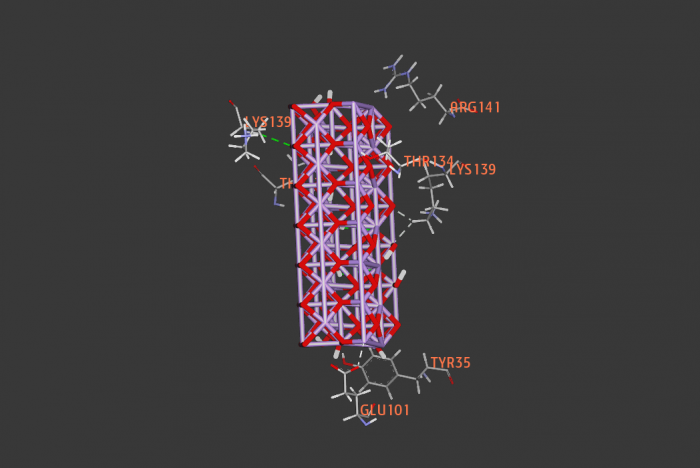
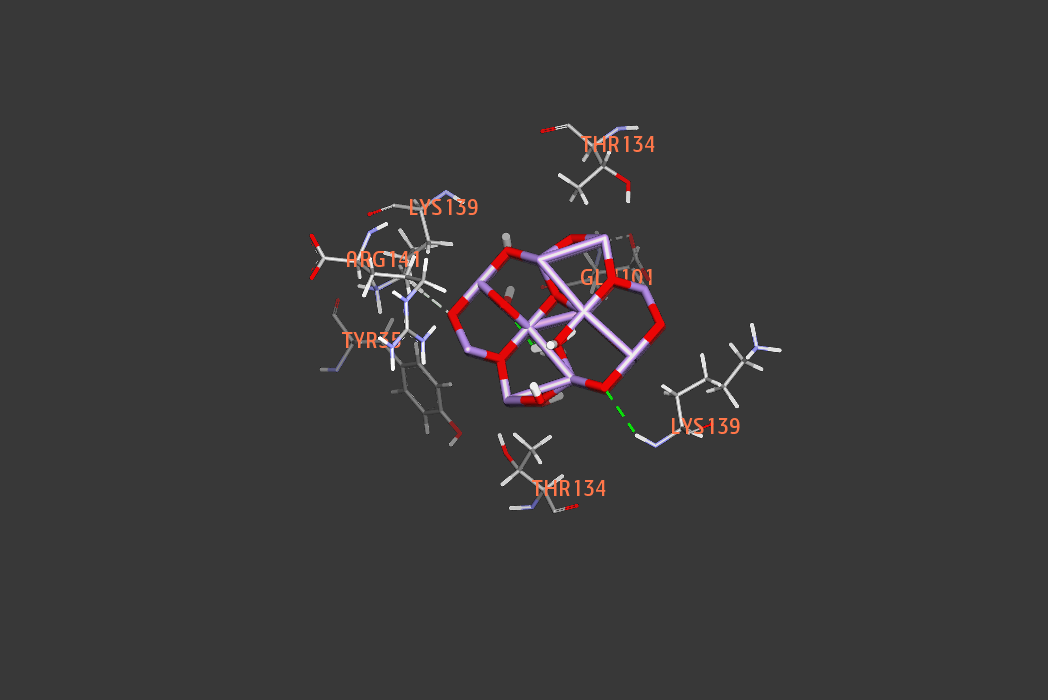
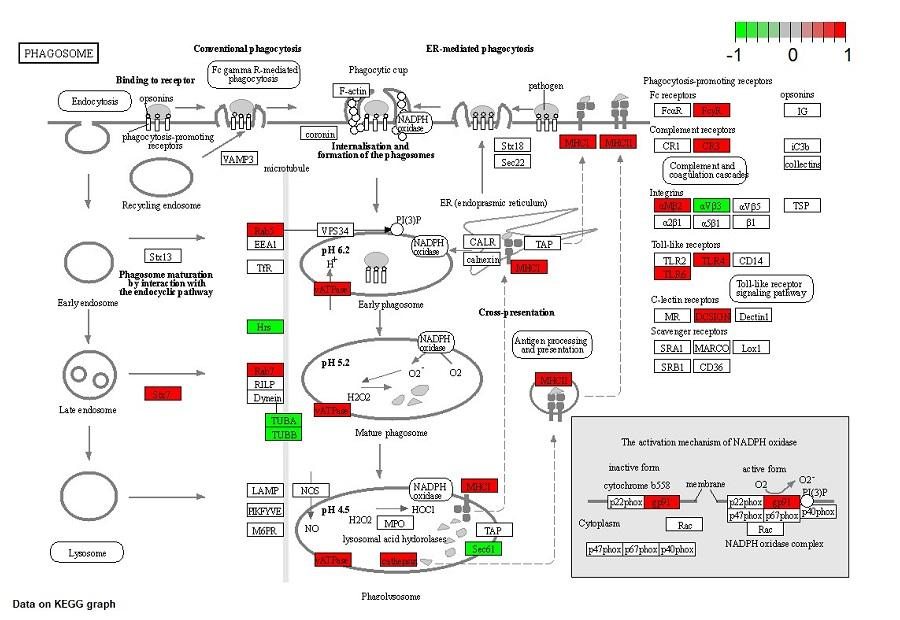
new inhibitors of Staphylococcus aureus sortase A -
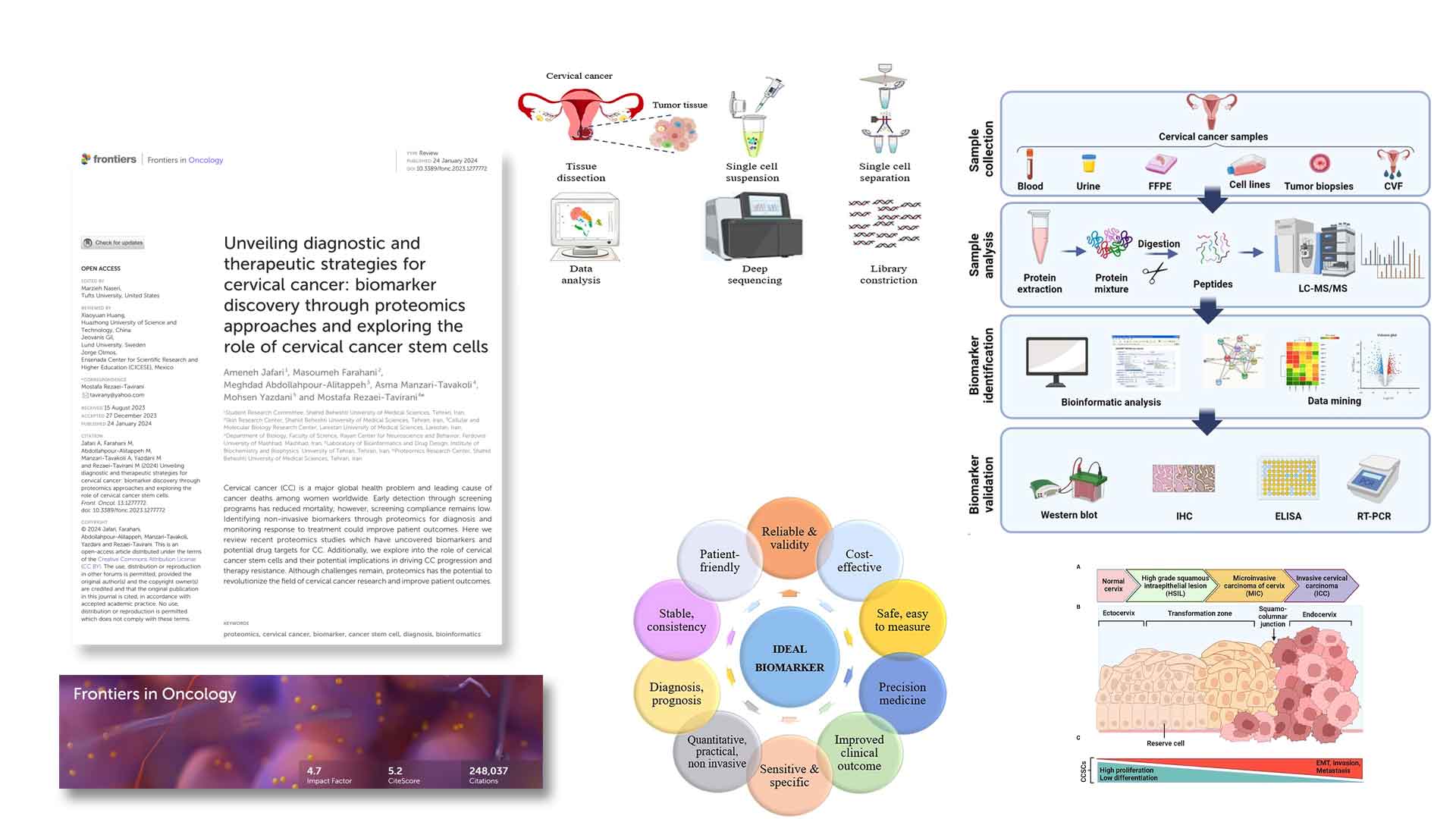
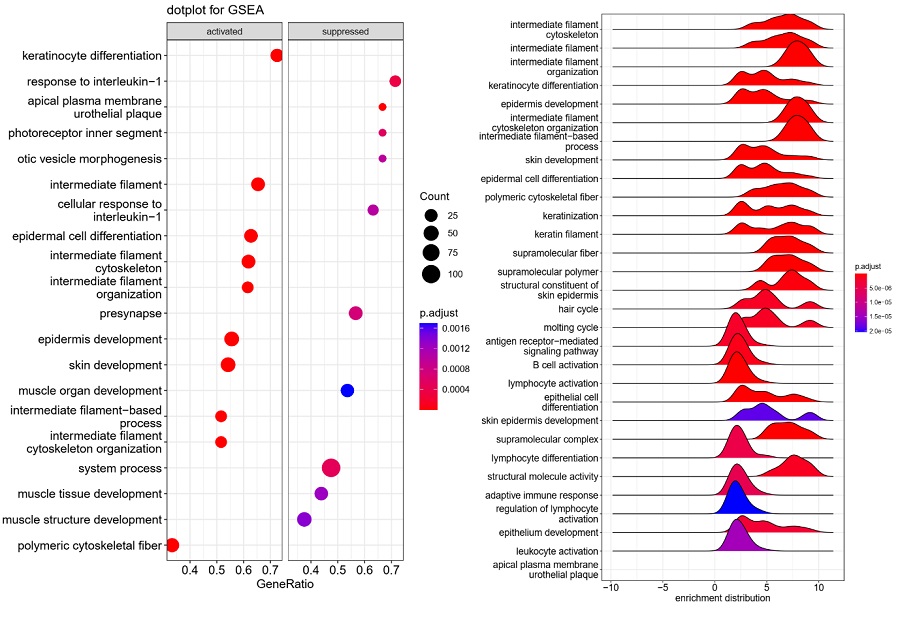
Unveiling diagnostic and therapeutic strategies for cervical cancer:
biomarker discovery through proteomics approaches ... -
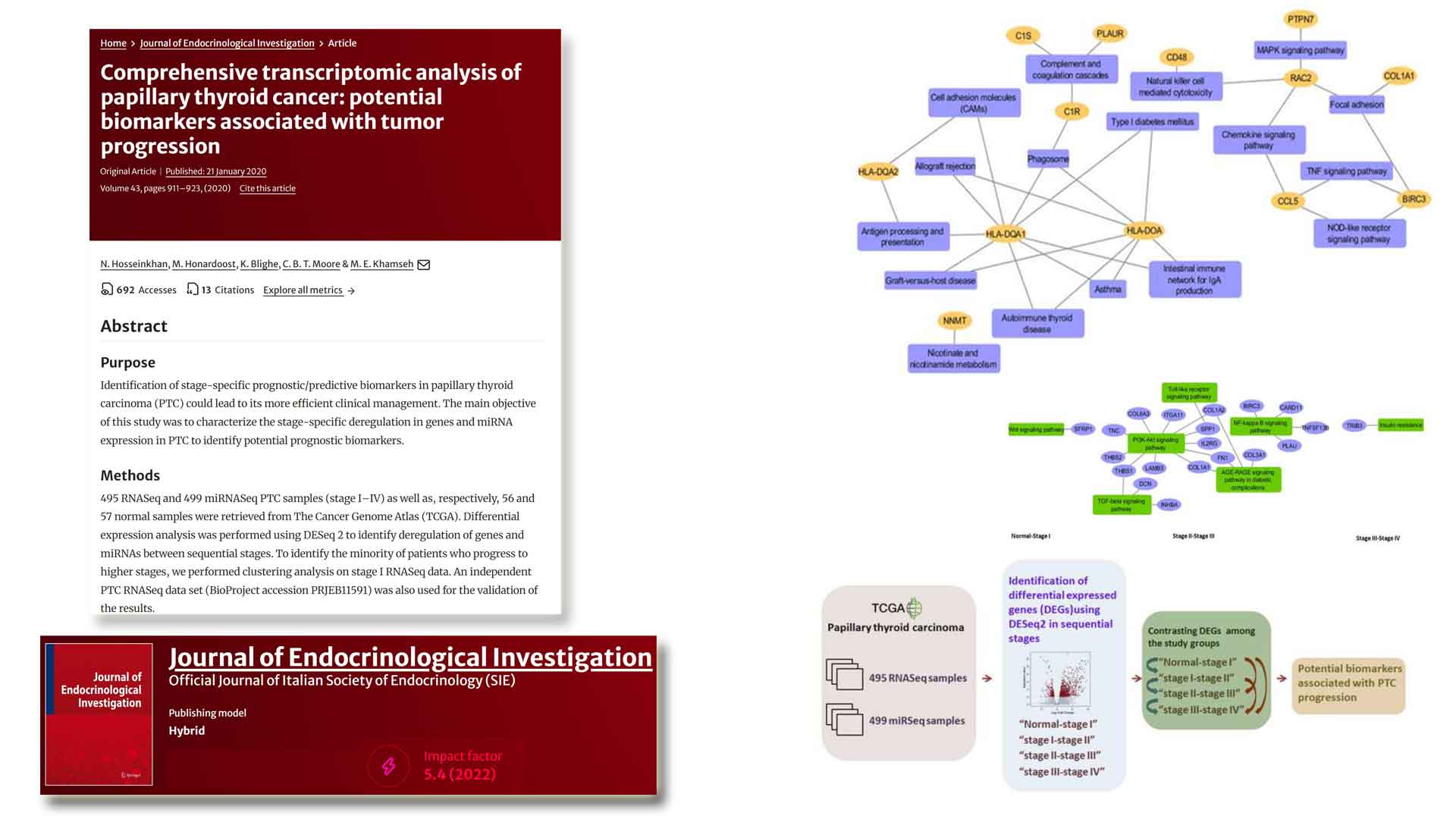
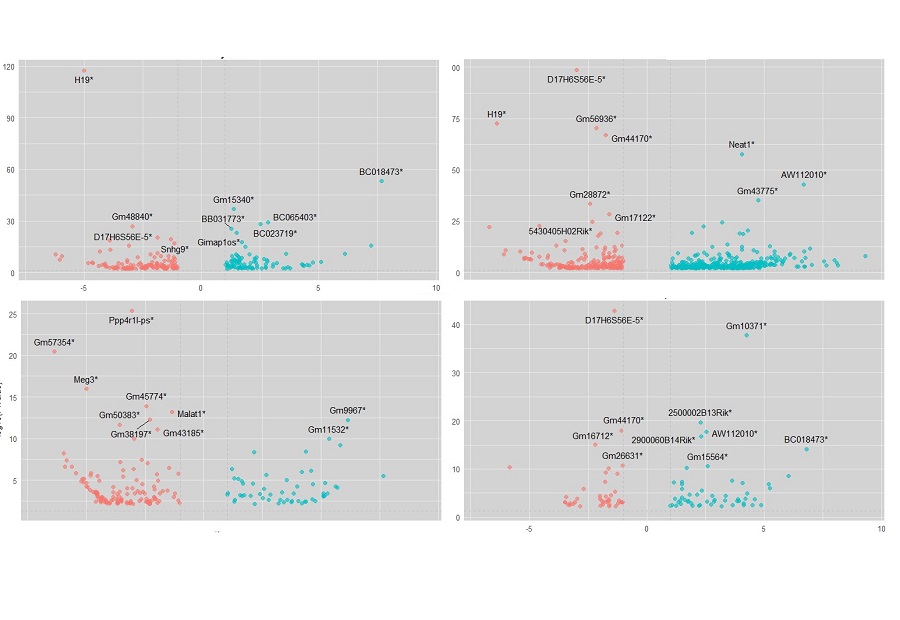
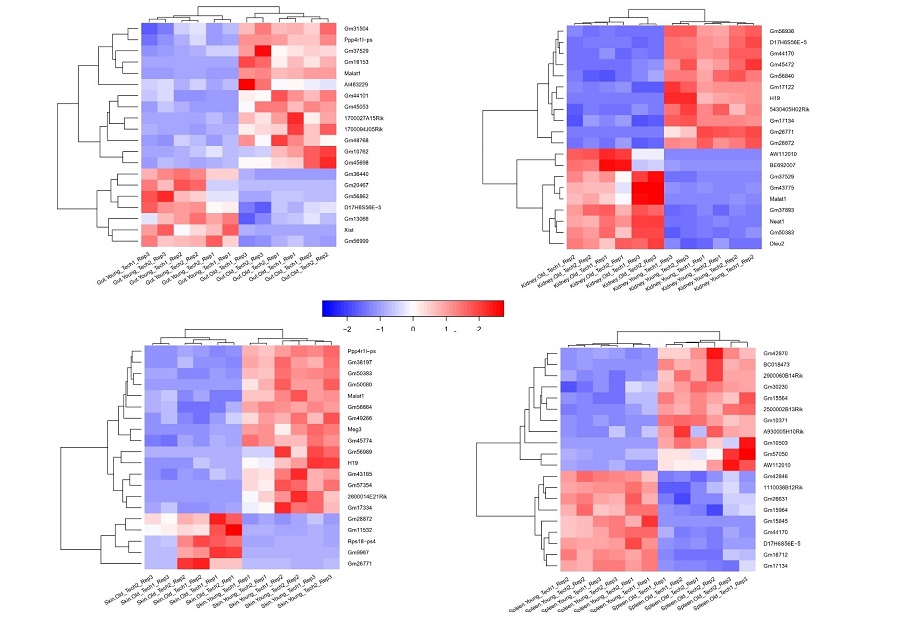
Transcriptomic analysis of papillary thyroid cancer:
potential biomarkers associated with tumor progression -
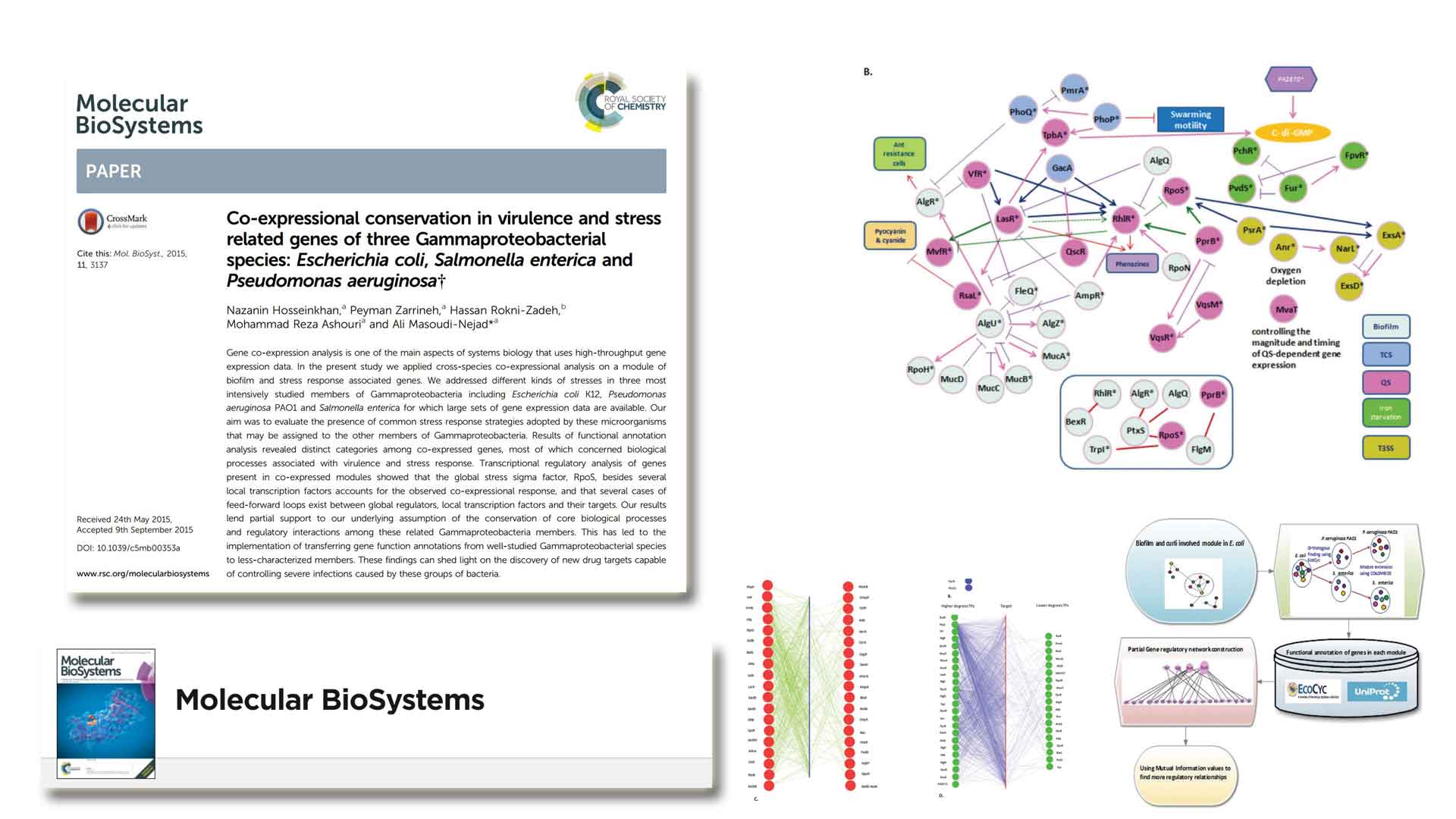
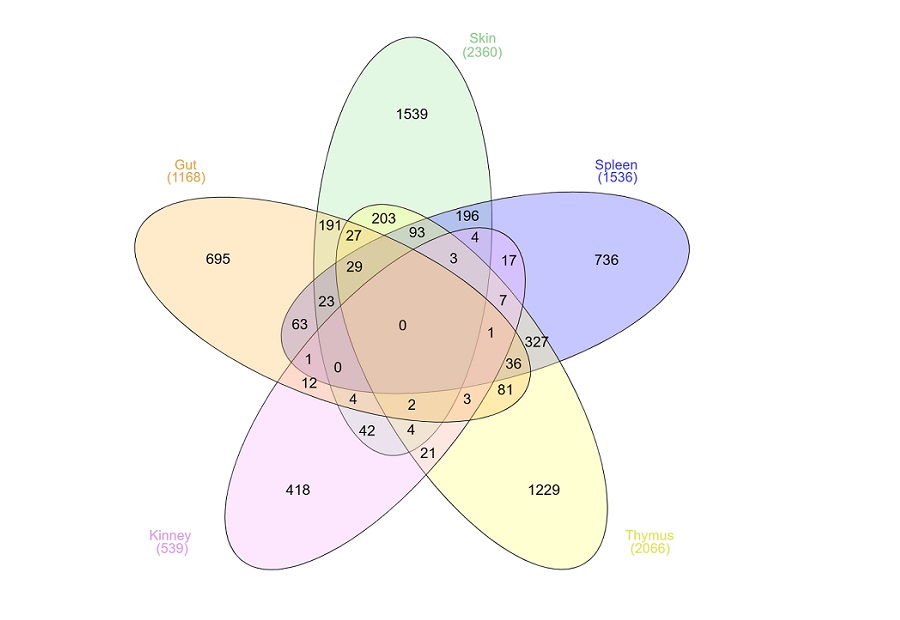
Co-expressional conservation in virulence and stress related genes of three Gammaproteobacterial species ...
- Rational approaches,
SARS-CoV-2/ACE2 PPI inhibitors - Drug repurposing approach,
FDA approved drugs against RdRp and 3CLpro. - Structure-based virtual screening & MD,
new inhibitors of Staphylococcus aureus sortase A - Unveiling diagnostic and therapeutic strategies for cervical cancer:
biomarker discovery through proteomics approaches ... - Transcriptomic analysis of papillary thyroid cancer:
potential biomarkers associated with tumor progression - Co-expressional conservation in virulence and stress related genes of three Gammaproteobacterial species ...